<<< Previous page | | Next page >>>
Molecular mechanisms of taste-avoidance learning
We are interested in revealing the mechanisms of behavioral plasticity and learning behavior. Caenorhabditis elegans is useful for the analysis of mechanisms at the molecular levels because it has simple neural circuits and genetic analysis is easily performed on this organism.
We found the associative learning in C. elegans as described below. C. elegans cultivated with food shows positive chemotaxis to NaCl, whereas C. elegans that experienced starvation with NaCl for a given length of time shows negative chemotaxis to NaCl. However, this change doesn’t occur by starvation without NaCl or presentation of NaCl in the presence of food. Therefore, this phenomenon is associative learning in which the animals remember the association between NaCl and starvation. We call this learning “taste-avoidance learning”.
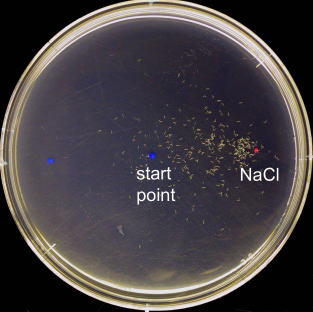
When C. elegans is placed at the center of the plate with a NaCl concentration gradient, they are attracted to the high concentration side.
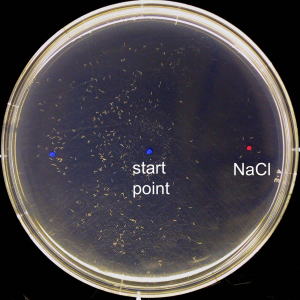
When C. elegans was placed at the center of the plate with a NaCl concentration gradient, it avoids the high concentration side.
The molecules that act for taste-avoidance learning
Water-soluble molecules, such as NaCl, are sensed by the “ASE” sensory neurons. But what molecules function in ASE neurons and neurons connected to ASE neurons when C. elegans performs associative learning? To answer this question, we’re searching for mutants that are unable to perform taste-avoidance learning properly.
So far, we have found that mutants of the insulin-like signaling pathway and calsyntenin homolog CASY-1 have defects in learning. These molecules have essential function in learning.
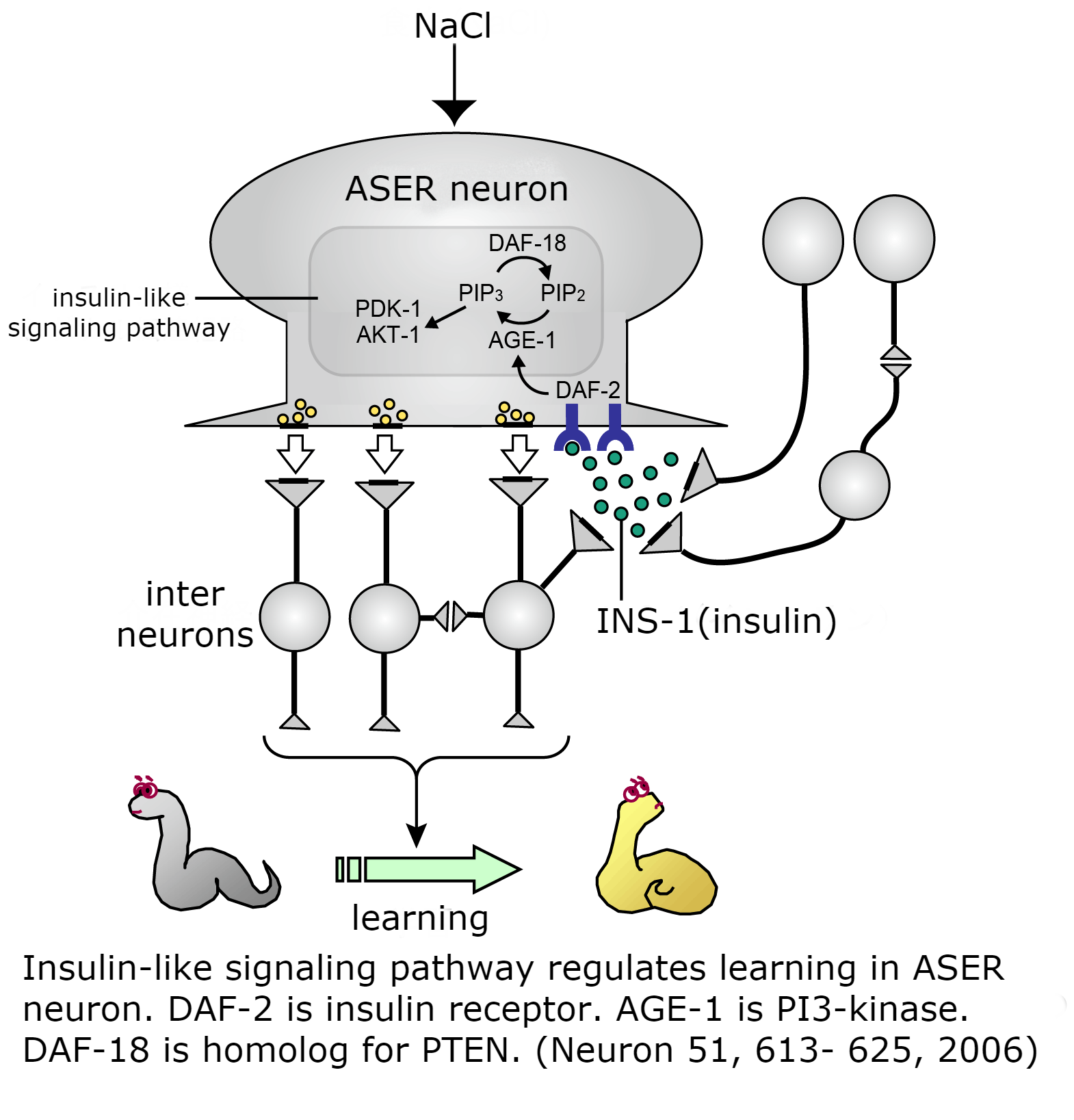
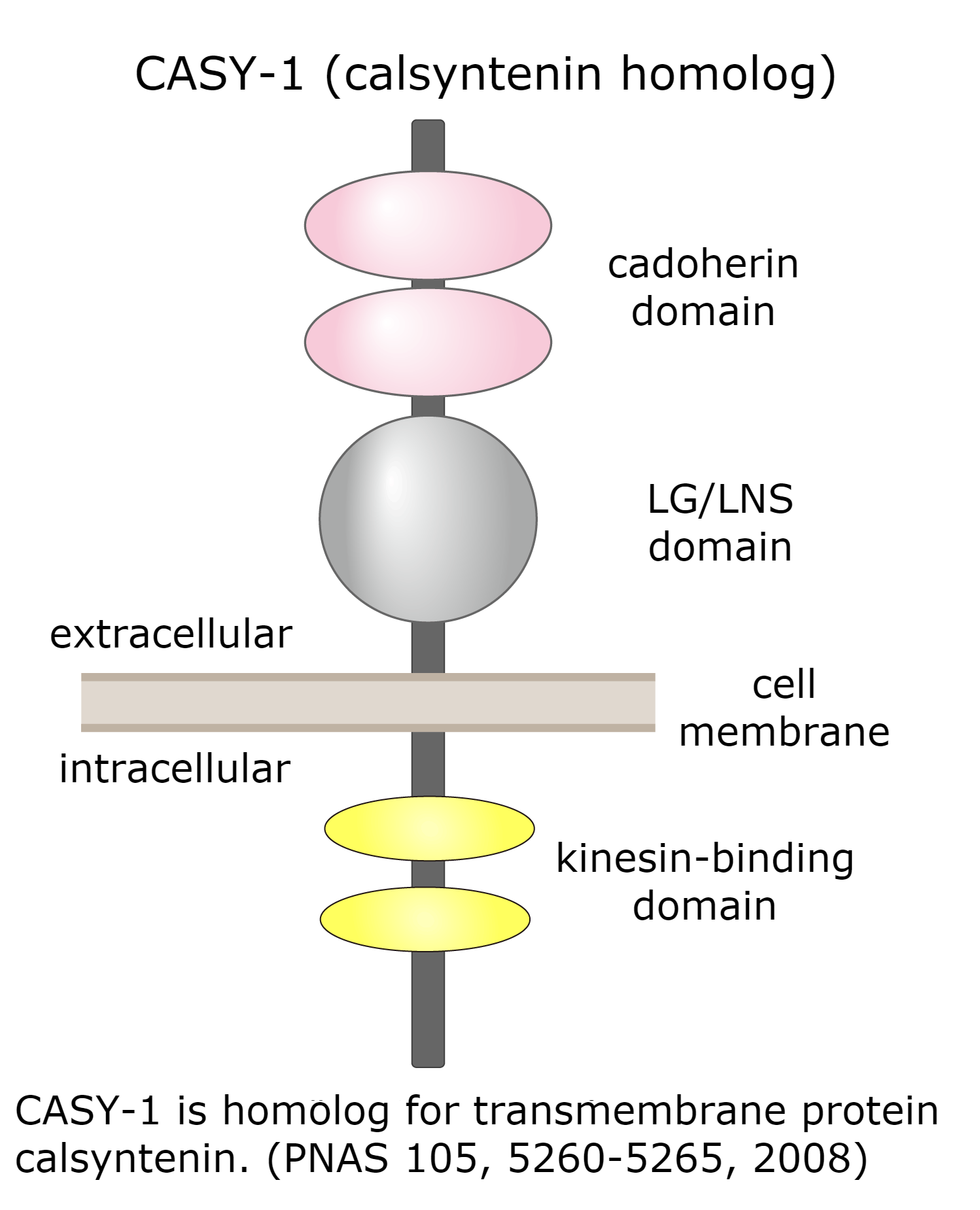
Identification of the cell in which the molecules act for controlling learning
PI 3-kinase AGE-1 which functions in the insulin-like signaling pathway and calsyntenin CASY-1 regulate learning in the neuron called ASER. CASY-1 is not a component of the signaling pathway, but needed for transportation of insulin receptor to synaptic site of the neuron where the signaling pathway act. ASER neuron is a member of the ASE sensory neurons described above. ASE sensory neurons are composed of ASEL (the left side ASE neuron) and ASER (the right side ASE neuron), which are asymmetrical and similar in structure but have differences in important functions. Our experimental data has revealed that ASEL and ASER act differently in learning.
INS-1 functioning in learning
C. elegans has 40 insulin-like molecules. Of these, INS-1 has an important function in taste-avoidance learning. INS-1 is generated in the intestine and neurons but the neuronal INS-1 is important for controlling learning. INS-1 in these neurons localizes at presynaptic sites and is secreted from the site. INS-1 functions like a neuropeptide.
Insulin and calsyntenin act in brains of higher animals
Although much remains unknown as to how brain works in mammals including human being, insulin and calsyntenin also act in the brain of mammals. These molecules are involved in memory and dementia. Research using C. elegans may lead to elucidation of their roles.
The insulin-like signaling pathway and the DAG/PKC pathway
Switching of salt chemotaxis by learning is reversible. Switching from attraction to avoidance of salt is caused by activation of the insulin-like signaling pathway in ASER gustatory neuron. For further understanding of this learning, we studied the regulation of switching from avoidance to attraction to salt and found that activation of the DAG/PKC pathway is necessary for attraction to salt. In this pathway, PLC (phospholipase C) generates DAG (diacylglycerol) and IP3 from the phosphoinositide PI(4,5)P2 that exist in cell membrane. DAG activates TTX-4/nPKC and promotes attraction to salt. Thus, it’s thought that the insulin-like signaling pathway and the DAG/PKC pathway regulate switching antagonistically. Furthermore, we identified the genes, pitp-1/phosphatidylinositol transfer protein and plc-1/phospholipase C, which are essential for control of salt chemotaxis learning. These genes are necessary for generation of DAG at presynaptic sites of ASER neuron, and it was found that these promote attraction to salt. By analyzing mutants of these genes, it was suggested that output of regulation of attraction and avoidance partly overlap. That forms foundation of understanding the mechanism of switching in salt chemotaxis learning.